Researchers identify new genetic cause of severe immune disorder
 Thursday, April 23, 2020 at 7:01PM
Thursday, April 23, 2020 at 7:01PM Severe congenital neutropenia leaves young patients to contract infection after infection, leading to life-threatening situations. A team of Leuven scientists has identified a novel genetic mutation, pointing to a new causative mechanism for this severe immune disorder.
The story starts with patient Jane Doe, now 19 years old, but diagnosed with severe congenital neutropenia when she was just 2 years old. By that time, she had already suffered an ear abscess, recurring ear infections, bronchitis, sinusitis, tonsillitis and several gum infections.
 After yet another infection, this time of her intestine, a detailed investigation revealed a striking shortage of neutrophils, white blood cells that are recruited as first-responders to the site of injury or infection within our body. Having an abnormally low concentration of neutrophils in the blood is referred to as neutropenia. When it is severe and present from birth (congenital), that is where the diagnosis of severe congenital neutropenia comes in.
After yet another infection, this time of her intestine, a detailed investigation revealed a striking shortage of neutrophils, white blood cells that are recruited as first-responders to the site of injury or infection within our body. Having an abnormally low concentration of neutrophils in the blood is referred to as neutropenia. When it is severe and present from birth (congenital), that is where the diagnosis of severe congenital neutropenia comes in.
“Severe congenital neutropenia is very scary, because these kids develop serious infections that can be lethal for infants,” explains Erika Van Nieuwenhove. “As if that’s not enough, they are also at increased risk for other conditions such as leukemia.”
Van Nieuwenhove is both an MD and PhD, who combines clinical work in the university hospital with Carine Wouters, with research at VIB and KU Leuven under the guidance of Adrian Liston and Stephanie Humblet-Baron.
Together with John Barber and several other colleagues, she set out to understand why Jane Doe developed SCN in the first place. Van Nieuwenhove: “For up to 50% of severe congenital neutropenia patients, we have no clue what causes the disease. It was the same for our patient, whose parents are both healthy.”
A new mutation in a familiar gene
After Jane Doe tested negative for mutations in all the genes with known ties to neutropenia, the researchers performed whole exome sequencing, probing every gene in the DNA, to trace back the genetic defect underlying the disorder.
“We identified a new mutation in a gene called SEC61A1, which encodes one of three subunits of the Sec61 complex. This molecular complex plays a crucial role in both protein transport and in maintaining the calcium balance of the cell,” explains Humblet-Baron. “Our experiments revealed that the genetic defect led to both a lower expression and a reduced efficacy of the SEC61A1 protein, and that these quantitative and qualitative defects in turn disturb neutrophil differentiation and maturation.”
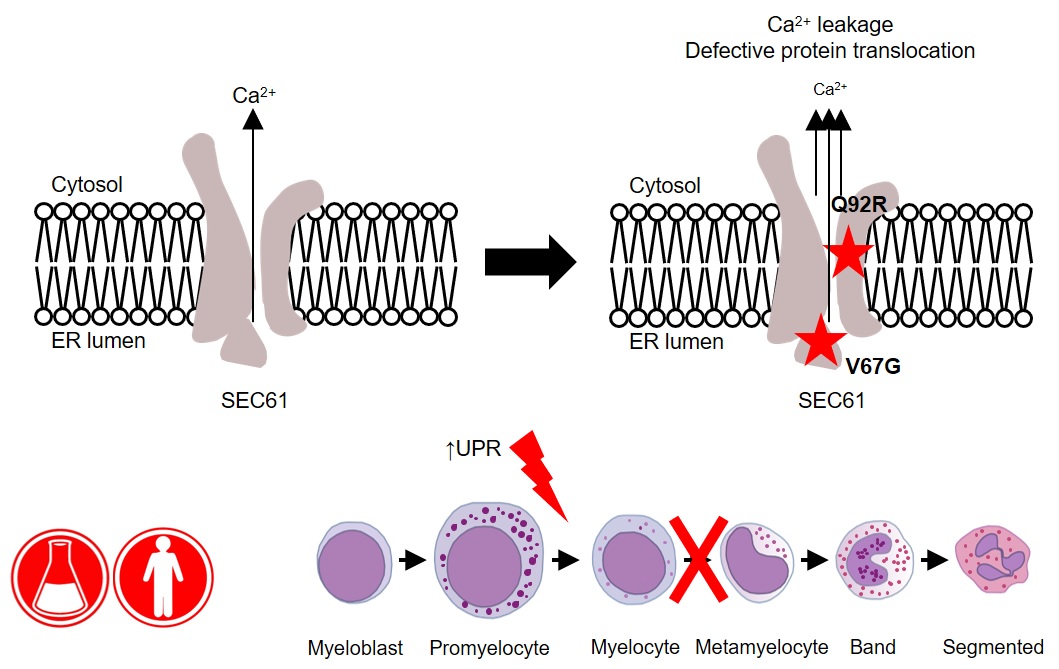
Interestingly, SEC61A1 has recently been picked up in other studies that were not focused on neutropenia. Different mutations in the same gene were reported in two families with a rare kidney disease and in two additional families with an antibody deficiency.
“The fact that there are different mutations in the same gene indicates there may be overlapping mechanisms among the different disorders. With the low number of currently known patients, it is still too early to predict which mutations can lead to which symptoms,” explains Liston.
“What’s clear from our findings is that SEC61A1 mutations can also cause severe congenital neutropenia. Considering this gene’s link with other disorders, the clinical implications of our work reach far beyond the patient with whom it all started here in Leuven.”
—
Read the original paper: Defective SEC61α1 underlies a novel cause of autosomal dominant severe congenital neutropenia. Van Nieuwenhove et al. JACI 2020







